Introduction
The primary deliverable of the project is to set up a cloud pilot for bridging the collaborative needs of the various organizations of ICTBiomed community. The architecture presented in this document is a reference for implementation of a cloud-based infrastructure built on open standards and new technologies to provide a scalable, flexible and dependable framework for optimized delivery of data-intensive software services. This document focuses on identifying requirements for an infrastructure for the reliable and effective high level architecture for a system that can satisfy collaborative requirements. The goal of this document is to set the foundation on which the components that make up the Cloud infrastructure will be developed.
Requirement of collaborative Cloud environment
For the past many years ICTBiomed consortium has been bringing better ways to work with many solutions in the area of Biomedical research. The consortium is looking forward to further improve the role of life sciences supercomputing centers in India, Poland, Sweden and USA, working together. The consortium, has created a science-led next generation cyber- infrastructure capability with a wide range of biomedical research projects. The emergence of cloud computing has changed every aspect of the collaboration and provided a ground change for research. The ICTBiomed consortium has planned to create a pilot cloud to provide these capabilities to fulfill the basic research requirements of researchers across the globe. The challenge currently exists that there are multiple locations and competing views of what cloud computing requirements. Cloud standards are still in flux. Yet, there is a consistent view that cloud computing will definitely be a ground change for collaborative research. These changes will go beyond the technology aspect and into how various research organizations work together. It will require the need for a standard agreement among the various peers having their own technology skill sets, governance and organizational requirements and encompass a common view of consortium cloud. Cloud platforms can enable life sciences community to work more effectively with many multidisciplinary experts. Many scientists today find it a challenge to manage and analyze their data. There is a growing gap between the ability of modern scientific instruments to produce data and the ability of scientists and most research groups to manage, analyze, and share the data that is produced. Indeed, as the size of data-sets grows from MB to GB and TB, many scientists are finding it difficult just to transport their data from the instrument where it is produced to the facility where it is analyzed, and from the facility where it is produced.
Objective of Global ICTBiomed Cloud Pilot
* Unified interface to manage Cloud resources distributed in different locations like PSNC (Polland), Chalmers (Sweden), OHSL (USA) and C-DAC (India).
* Common Cloud architecture agreement among peers.
* Enable VM migration between clouds.
* Test multi-zone cloud infrastructures.
* Test resources sharing.
* Scientific use-cases for multicloud projects.
Proposed Reference Architecture
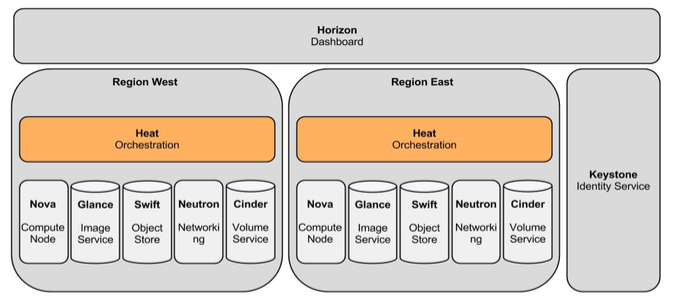
The proposed reference architecture consists of a common agreement of Cloud middle-ware software which should be installed at all the locations as a part of the consortium. The various locations should be connected by a high speed link capable of transfer of the required data. All the location are identical except the entry gateway location which has an additional security and authentication mechanism for the user accessing the cloud. The model works on resource sharing mechanism and all the resources are transparent to the user. The location whose resources are available can be accessed by the user without knowing the actual site location. Another mechanism can be adopted where the various partner locations can be represented as zones and the user can access the desired zone as per the proximity to the location.
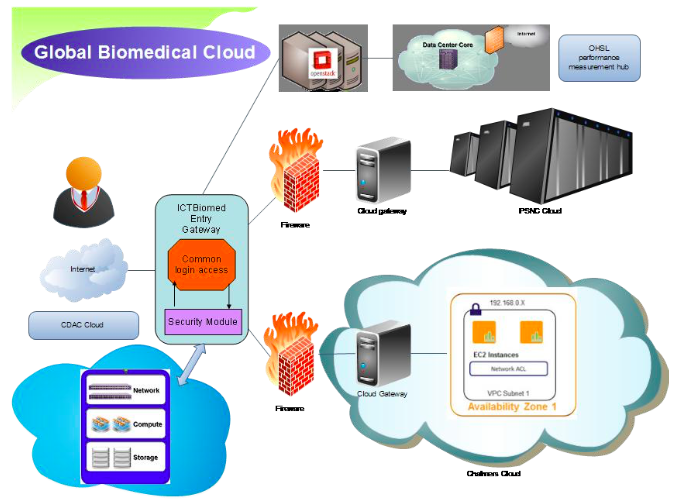
Fig 1. Proposed reference Architecture
The various components of the architecture are explained below:
1) Cloud Middleware : OpenStack is quickly becoming the de-facto standard for Open Cloud platforms. The installation can be performed very easily at all the sites remotely or by local system administrators.
The components of OpenStack are:
* Dashboard (“Horizon”) provides a web front end to the other OpenStack services
* Compute (“Nova”) stores and retrieves virtual disks (“images”) and associated metadata in Image (“Glance”)
* Network (“Quantum”) provides virtual networking for Compute.
* Block Storage (“Cinder”) provides storage volumes for Compute.
* Image (“Glance”) can store the actual virtual disk files in the Object Store(“Swift”)
* All the services authenticate with Identity (“Keystone”)
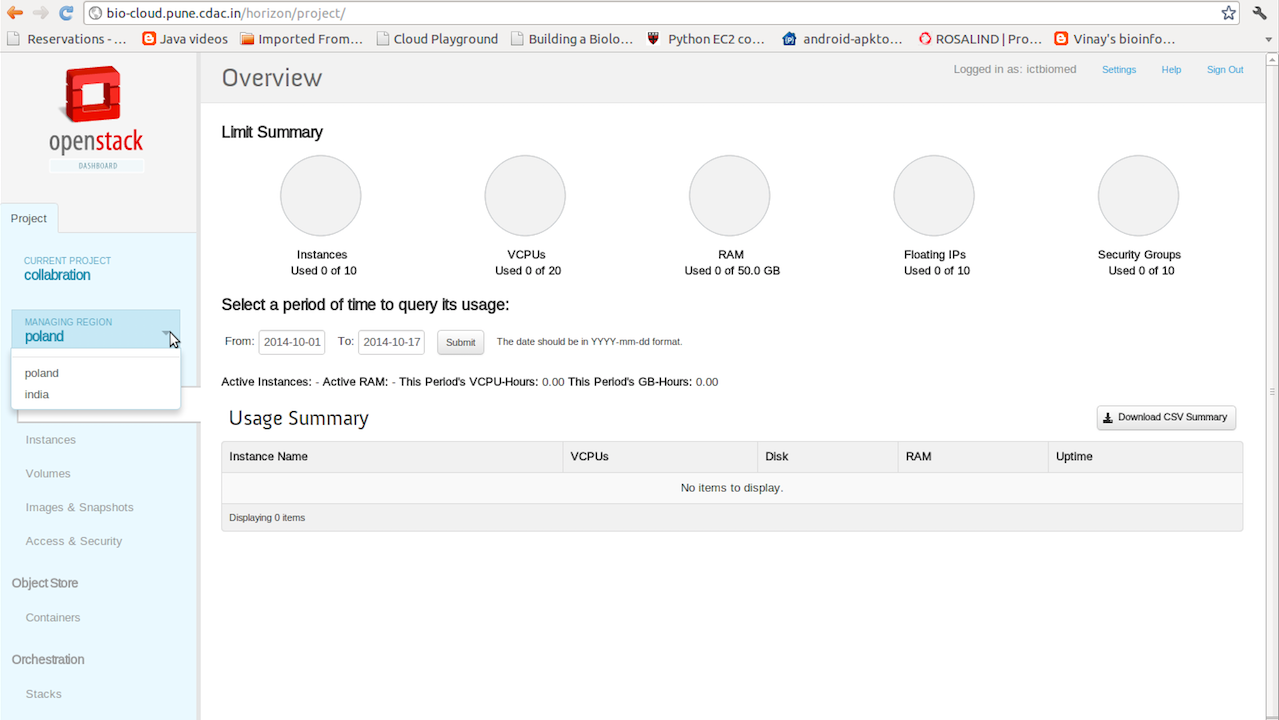
Fig 2. Dashboard for Cloud Administration
2) Zones : Openstack cloud can be zoned from top to down levels, into Regions, Availability Zones and Host Aggregates :
* Region : Each Region has its own full Openstack deployment, including its own API endpoints, networks and compute resources. Different Regions share one set of Keystone and Horizon to provide access control and Web portal.
* Availability Zone : Inside a Region, compute nodes can be logically grouped into Availability Zones, when launching new VM instance, we can specify Availability Zone or even a specific node in a Availability Zone to run the VM instance.
* Host Aggregate : Besides Availability Zone, compute nodes can also be logically grouped into Host Aggregates Host Aggregates have meta-data to tag groups of compute nodes, e.g. we can group nodes with SSD disk to one Host Aggregate, and nodes with 10 GB NICs to another Host Aggregate. One compute node can be put into Host Aggregate and Availability Zone and the same time, Host Aggregate has no conflict with Availability Zone. One compute node can be put into more than one Host Aggregates Host Aggregate is visible only to admin, for end-user, it’s exposed by creating customized flavor with Host Aggregate meta-data linked.
Fig 3. The Cloud zones of OpenStack
The cloud can be divided into four regions:
A) C-DAC region : This region has the entry gateway. The entry gateway is the authentication and credential management server which authenticate a genuine user. In the proposed architecture it is shown located at C-DAC. It can be located at any of the location where the cloud master node is installed. This region also has its own network, compute and storage nodes which can be accessed as an OpenStack node.
B) PSNC region: This region is having a vast pool of resources which can be effectively utilized by the community through the OpenStack cloud availability platform.
C) Chalmers region: This region has technologies and data crunching algorithms which can be provide as SaaS to the community. The pipelines intelligently uses the rich biomedical data resources.
D) OHSL/Notre Dame : OHSL/Notre Dame is a very important region which measures the heartbeat of the full cloud infrastructure. This node can be used as a performance measuring hub for monitoring the effective utilization of the cloud resources.
Regions segregate an entire cloud and result in running separate openstack deployments. Region can be thought of as different datacenters and have complete installation openstack. This deployment has central authentication & dashboard.
So instead of having two keystone service( OpenStack Identity Service) means two databases of user, we will be having single keystone & dashboard.
Defining service endpoints:
Each of the services in openstack environment runs on a particular URL and port—these are the endpoint addresses for services. When a client communicates with our OpenStack environment that runs OpenStack
Identity Service, it is this service that returns the endpoint URLs, which the user can then use in an OpenStack environment. To enable this feature, we must define URL endpoints separately for each region. In a cloud environment, though, we can define multiple regions. Regions can be thought of as different datacenters, which would imply that they would have different URLs or IP addresses. In keystone database we have two tables service & endpoint. So in multi region scenario we will be having only one service entry for every deployment.
And n entries in endpoint table for services running on n Datacenters.
#keystone service-create —name nova —type compute —description ‘OpenStack Compute Service’
#keystone endpoint-create —region india —service_id $ID —publicurl $PUBLIC —adminurl $ADMIN —internalurl $INTERNAL
#keystone endpoint-create —region poland —service_id $ID —publicurl $PUBLIC —adminurl $ADMIN —internalurl $INTERNAL
Note – service_id must be same if configurating same service (i.e same for nova ) & is output of keystone service-create command. And for central authentication, any query to keystone service from any region must return same keystone endpoint.So we will doing n entries in endpoint table for n regions.
Fig 4. Regions using OpenStack cloud
3) Cloud Gateway: Cloud gateway are one more level of security mechanism maintained by every site which is a part of the global cloud. It is located at every participating site behind the firewall.
4) Local Cloud Site: The local Cloud site is a location where the actual resources are available. These resources can be huge supercomputing resources like PSNC or it can be scientific data useful for analysis. Every location can be shown as a availability zone which can be accessed by the user based on their requirements.
Performance Benchmark
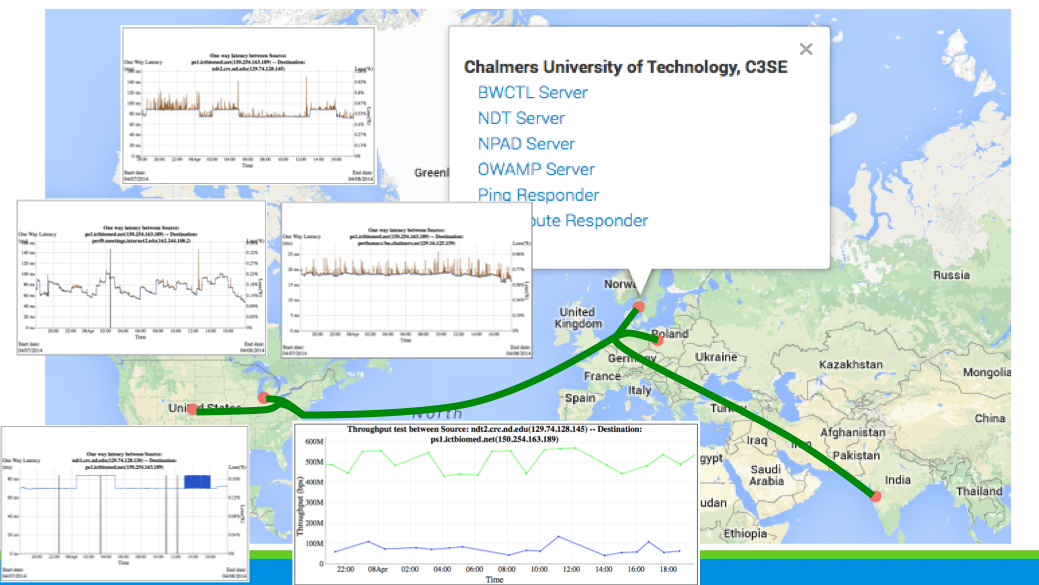
We will use perfSONAR for network performance monitoring. perfSONAR is an infrastructure for network performance monitoring, making it easier to solve end-to-end performance problems on paths crossing several networks. It contains a set of services delivering performance measurements in a federated environment. These services act as an intermediate layer, between the performance measurement tools and the diagnostic or visualization applications. This layer is aimed at making and exchanging performance measurements between networks, using well-defined protocols. perfSONAR is well suitable for a consortium of organizations who seek to build network performance middleware that is interoperable across multiple networks and useful for intra- and inter-network analysis. One of the main goals is to make it easier to solve end-to-end performance problems on paths crossing several networks.
perfSONAR-PS is a collection of software packages used to monitor and measure networks. There are two major ways this can be integrated into a networking environment.The goal of the perfSONAR-PS effort is to demystify the process of network monitoring.
The plan is to use All-in-one monitoring solution, in the form of the pS Performance Toolkit installed via CD or USB Key. This is the suitable choice if the goal is to have a dedicated monitoring host on the network.
Download Link: http://software.internet2.edu/pS-Performance_Toolkit/
The perfSONAR-PS virtual cloud image will be used on OpenStack cloud middleware.
Plan of Action:
*Identify Technical contact points at the four locations. *Setup a Global Cloud Test Bed which will involve identifying a server at each of the three locations on which the cloud middleware will be installed. *OHSL terminal as a network performance measurement node to demonstrate access of the Global Biomedical cloud. *Testing the network speeds across the three locations. *Making basic cloud test runs. *Plugging in an application pipeline to run across the three locations
Configurations of participating nodes
As a part of Proof of Concept(POC) phase we can include only a single node with good public network connectivity at every location (zones). The nodes should be capable enough in terms of processor and memory to support the loading of virtual OS images like CloudBioLinux. As a template following confirurations should be followed:
1) Atleast 8 processing cores 2) 16 GB of memory 3) 1 TB HDD capacity 4) Hardware virtualization support 5) Four NIC ports
Challenges
We have to overcome the geographic distance between participants, lack of central authority, and heterogeneous multiple technologies, physically separated from one another by long distances. Hurdles in the deployment of an infrastructure, including the lack of a hierarchical organization and heterogeneous approaches to the implementation of core middle-ware services. The Cloud Setup infrastructure presents a unique opportunity for the collaborators to peer with one another and form a collaborative environment.
Conclusion
We will demonstrate how this Global Biomedical Cloud pilot where an object data model, computational storage, content-centric access, comprehensive data interoperability, and security guarantees play a central role supports new emerging biomedical services. In doing so, we aim to achieve significant and quantifiable improvements in solving the problems which can not be solved by an individual organization but can be solved by collaborative expertise joined together. Setting a Global Cloud infrastructure have enormous capabilities of progressive research.